Chloroflexi
Detection of the group of Chloroflexi filaments and specific
identification of Eikelboom Type 1851 in sludge samples
Item Number: 01130020
Lab analysis: detection of filamentous bacteria
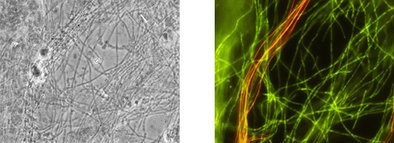
The group of Chloroflexi (formerly: green, non-sulfur bacteria) are mostly filamentous bacteria. Their members show a high morphological similarity while diverging highly on genetic level. The Gram-variable bacteria mostly are non-cultivable, however, their numbers and related problems increased enormously over the last decade. In case of massive growth they can create severe sludge settling problems.
Chloroflexi can be found not only in aerobic wastewater treatment plants but also in anaerobic pellet sludge where they are part of the native populations. Especially in anaerobic sludges of the paper industry, the group of Chloroflexi quantitatively plays a major role within the biocenosis. Thus, some representatives seem to support hydrolysis while others exert a negative influence on pellet stability which can lead to pellet disintegration and subsequent pellet loss.
With the VIT® gene probe technology, the important group of Chloroflexi filaments and of Eikelboom Type 1851 are identified and quantified in sludge samples. Thus, before problems arise, important conclusions regarding population composition and -shifts can be made. Use VIT® Chloroflexi rapid analysis as early warning system. Thereby, sludge degeneration phenomenons may be prevented and population shifts leading to negative consequences for plant performance may be identified in due time.
The results are displayed according to the VIT® key. The report contains a representative photo documentation that visualizes the Chloroflexi filaments in the wastewater sample.
Qualitative or quantitative detection?
Quantitative detection
Which method is applied?
The reliable VIT® gene probe technology.
Which requirements are important?
Please contact us for further details regarding your samples.
Choose Filaments PLUS for a complete profile of filamentous bacteria in your plant or discover our Premium population analysis.
Your Advantages
specific identification of unculativable Chloroflexi
specific identification of Eikelboom Type 1851
quantification
detection of living bacteria exclusively
early detection of population shifts
prevention of sludge degeneracy phenomena
applied for the prevention of disturbances