Premium population analysis
Generation of a detailed and quantitative profile of all
bacteria populations in the sample
Item Number: 01120001
Lab analysis - comprehensive bacteria profile of wastewater samples
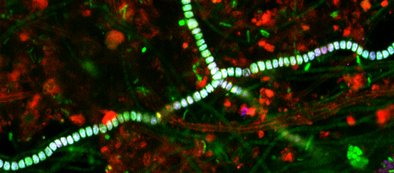
Establishment of a cultivation-independent population profile of all bacteria in a wastewater sample. Analysis is carried out using a selection of gene probes which differentiate 95-98% of all bacteria in wastewater samples according to their phylogenetic affiliation to bacterial main groups. All populations are specifically quantified in relation to the overall viable bacteria flora. Evaluation is conducted via microscope qualitatively/quantitatively.
The following 10 bacteria main groups are analyzed: Alpha-, Beta-, Gamma- and Deltaproteobacteria, Actinobacteria, Firmicutes, Cytophaga-Flexibacter-Subphylum, Planctomycetes, Chloroflexi, Nitrospirae.
Additionally, special important groups are analysed as well:
- filamentous bacteria are identified and quantified (they are differentiated in problem-causing organisms (bulking sludge) and more floc-forming filamentous bacteria)
- nitrifiying bacteria are analyzed on the group level (AOB and NOB)
Furthermore, total cell counts and total viable counts (cells/g (ml)) complete the performed analysis.
Summarizing report contains a significant photo documentation that visualizes the different populations in the sample material.
Qualitative or quantitative detection?
Quantitative detection
Which method is applied?
The reliable VIT® gene probe technology.
Which requirements are important?
Please request our simple protocol for fixation of wastewater samples. Make sure that you fixate/stabilize your samples in the indicated manner before sending the samples to us.
Also available: the Main group population analysis and the Population profiling in anaerobic reactors.
Your Advantage
comprehensive profile of all bacteria populations, including filamentous bacteria and nitrifying bacteria
quantification of each population
detection of living bacteria exclusively